Enzyme
n., plural: enzymes
[ˈɛnzaɪm]
Definition: biomolecule that acts as a catalyst for a chemical reaction
Table of Contents
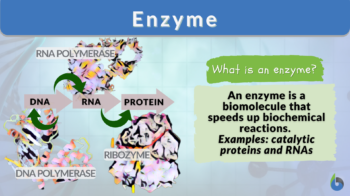
An enzyme is a biomolecule that can be synthesized biologically (naturally occurring) or through other processes (synthetically). Its main function is to act as a catalyst to speed up a reaction without itself being changed in the process.
Enzymes are commonly protein molecules with a characteristic sequence of amino acids that fold to produce a specific three-dimensional structure, which gives the molecule unique properties. Let’s read on to learn the enzyme definition, characteristics, types, and role in biological processes.
Enzyme Definition in Biology
An enzyme is a biomolecule that acts as a catalyst to speed up specific chemical reactions. Enzymes are either proteins or RNA molecules (ribozymes). Proteins are one of the major biomolecules; the others are carbohydrates (especially, polysaccharides), lipids, and nucleic acids. Enzymes that are proteins in nature are polymers of amino acids.
The amino acids are joined together by peptide bonds. The type and the sequence of amino acids in an enzyme protein structure are encoded by the DNA in the cell that produces them. While not all enzymes are proteins, not all proteins are enzymes as well.
Enzymes that are not proteinaceous in nature are exemplified by ribozymes. A ribozyme is an enzyme made of RNA rather than a protein. An example of a ribozyme is in the ribosome, which is a complex of protein and catalytic RNA units.
Etymology: from German “enzym”, from Medieval Greek “enzūmos”, meaning ‘leavened’.
Characteristics of Enzymes
Enzymes are often globular. They may occur singly or as a subunit in a complex. They are often larger than their substrates. While large relative to their substrate molecules, only a small portion of an enzyme is directly involved in catalysis.
The site involved in catalysis is referred to as the catalytic site. Another site in an enzyme structure is the binding site with which the substrate binds to or reacts with the enzyme.
The catalytic site and the binding site make up the enzyme’s active site.
The allosteric site of the enzyme refers to the site where another molecule can bind causing the enzyme to change its conformation, which then leads to its increase or decrease in its activity.
Biosynthesis
The production of enzymes by way of protein synthesis involves transcription and translation. Within the cells, an enzyme is generated by transcription and translation processes.
- Transcription is the process by which an mRNA template, encoding the sequence of amino acids in the form of a trinucleotide code, is transcribed from DNA to provide a template for translation.
- Translation is the process in which amino acids are linked together in a specific order according to the rules specified by the genetic code. It consists of four phases:
- (1) activation (the amino acid is covalently bonded to the tRNA)
- (2) initiation (the small subunit of the ribosome binds to 5′ end of mRNA with the help of initiation factors)
- (3) elongation (the next aminoacyl-tRNA in line binds to the ribosome along with GTP and an elongation factor)
- (4) termination (the A site of the ribosome faces a stop codon) – The newly formed proteinaceous structure will undergo subsequent processes, e.g., post-translational modification and folding.
Types
Enzymes are usually classified and named according to the reactions they catalyze. The International Union of Biochemistry and Molecular Biology have developed a nomenclature for enzymes, the EC numbers. They are as follows:
- EC 1 Oxidoreductases: catalyze oxidation/reduction reactions
- EC 2 Transferases: transfer a functional group (e.g. a methyl or phosphate group)
- EC 3 Hydrolases: catalyze the hydrolysis of various chemical bonds
- EC 4 Lyases: cleave various bonds by means other than hydrolysis and oxidation
- EC 5 Isomerases: catalyze isomerization changes within a single molecule
- EC 6 Ligases: join two molecules with covalent bonds
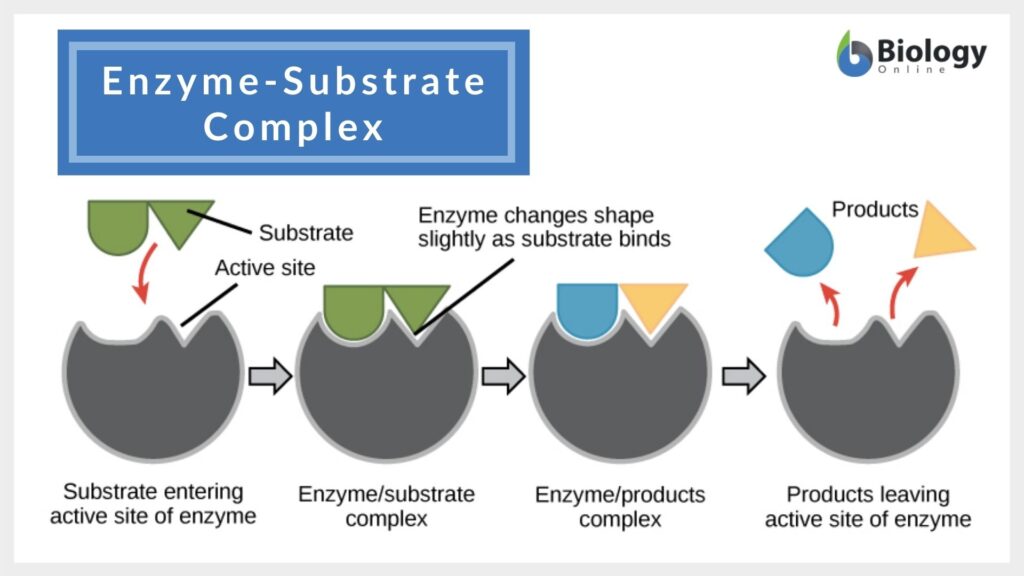
Enzyme-Substrate Interaction Models
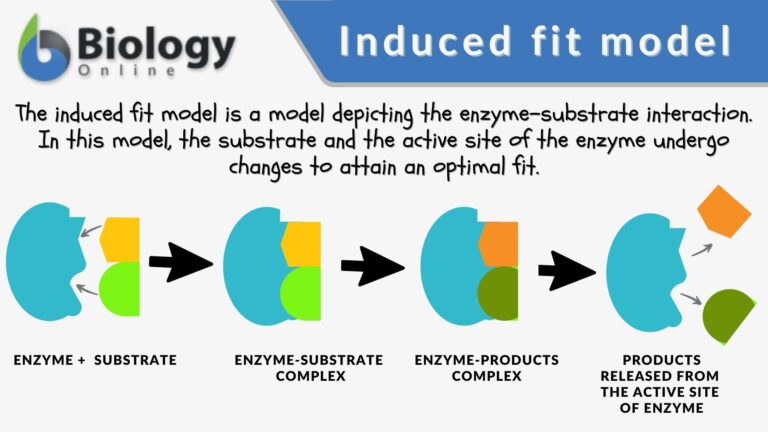
According to the induced fit model suggested by Daniel Koshland in 1958, the enzyme undergoes reshaping as it interacts with its substrate while the substrate may also change shape slightly so that they eventually fit into each other.
The enzyme speeds up a biological process by lowering the activation energy. It does so (1) by stabilizing the transition state, (2) by providing an alternative pathway, and/or (3) by destabilizing the substrate ground state.
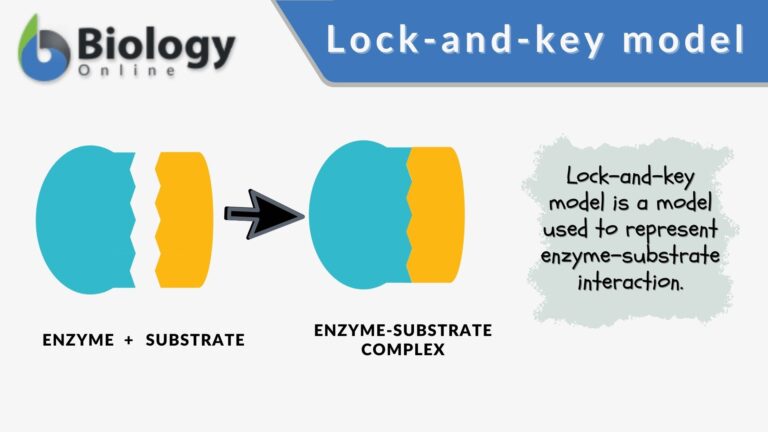
Another model that describes how the enzyme and substrate binding occurs is the lock and key model. In this model, high enzyme specificity is depicted.
The enzyme binds specifically only to a substrate that is an exact fit and only with such high specificity that the enzyme catalyzes the reaction. The interaction between the enzyme and the substrate is rather static and rigid as opposed to the more flexible interaction depicted by the induced fit model.
Mechanisms of Catalysis
Enzyme carries out catalysis together with the substrate in different ways:
- Covalent catalysis is where a temporary covalent bond forms between the enzyme and the substrate at the active site.
- Catalysis by approximation is where the enzyme orients multiple substrates by positioning them close together to improve the reaction rate.
- Acid-base catalysis involves the transfer of hydrogen ions (protons) from one molecule to another.
- Electrostatic catalysis employs electrostatic interaction with the substrate at the active site. Examples are catalysis that makes use of ionic, ionic-dipole, dipole-dipole, or hydrophobic interactions.
- Cofactor catalysis is a mechanism that employs cofactors, such as metals (e.g., iron, zinc, copper, manganese, etc.) and coenzymes.
Factors Affecting Enzyme Activity
Here are the typical factors that affect the catalytic activity of enzymes:
-
Biological factors (e.g., age and health)
There are various biological factors that could have an impact on enzyme activity. Examples of these factors are age and health condition.
The ability of the human body to naturally produce certain enzymes declines with age. Based on reports, there seems to be a decrease in the secretion of enzymes in the digestive system as the body ages. For instance, lactase production decreases with age, which is possibly one of the reasons lactose intolerance is experienced by the elderly. (NT Contributor, 2017)
The overall health condition of an individual also plays a part. Some individuals lack the genetic capacity to synthesize particular enzymes. For instance, high levels of liver enzymes in the blood could lead to excess liver enzyme production, which in turn, may be a sign of liver disease, such as hepatitis. (Cleveland Clinic, 2021)
Other enzyme imbalances associated with illnesses and metabolic disorders are:
-
- Exocrine pancreatic insufficiency is when the pancreas does not produce enough digestive enzymes (lipase, in particular) to break down complex molecules into smaller molecules, and so leads to poor digestion and absorption.
- Fabry disease is a genetic disorder that causes the body not to produce alpha-galactosidase A. This enzyme is essential in breaking down lipids (particularly, sphingolipids). Without it, globotriaosylceramide (a type of fat) builds up in the cells. This may potentially disrupt the normal functioning of the affected cells and organs, e.g., kidney and heart (Fabry Disease: MedlinePlus Genetics, 2022)
- Krabbe disease is another genetic disorder. It is caused by a genetic mutation that prevents the body from producing galactosylceramidase. This enzyme is essential in breaking down galactosylceramide and psychocine, which when in excess, are toxic to cells and therefore can damage myelin-forming cells.
-
Physical factors (e.g., temperature and pH)
Exposure to temperature and pH beyond optimal conditions leads to the enzyme’s inefficiency. For example, denaturation can occur when the temperature is exceedingly high. The enzyme loses its functional structure, such as the configuration of its protein chains.
The unique 3D structure of the enzyme determines how and where it will bind with the substrate. If damaged, the enzyme may not be able to bind with the substrate, and as a result, loses its catalytic capacity.
The enzyme reaction will be at its fullest when the enzymes are able to perform well, i.e., if the physical conditions, such as temperature and pH, are at optimal levels.
Note it!
The normal body temperature of adult humans ranges from 97.5°F to 98.9°F (36.4°C to 37.2°C). Therefore, most enzymes of the human body will work best within this range, particularly at around 98.6°F (37°C). That is why high fever (above 41°C) could impair bodily functions as some proteins, including those enzymes that are heat-sensitive, may become dysfunctional due to denaturation.
-
Chemical factors (e.g., chemical concentrations)
Chemical concentrations are also important factors in enzyme reactions. It may be in the form of enzyme concentration, substrate concentration, and the presence of inhibitors and/or activators. They affect the general catalytic ability of enzymes.
When there are a few enzymes available this could end up in slow biochemical reactions. But when the enzyme concentrations are high, this could, then, result in heightened catalytic reactions, provided that there are similarly a great number of substrates present at that time. When all substrates are used up, any further addition of enzymes will no longer speed up the reaction.
If most enzymes are unbound, then, adding more substrates to the reaction could increase the rate of reaction but also to a certain point. Adding more substrates while all enzymes are bound will not increase the rate of reaction any further. The enzymes have to be released first after being used in a reaction before they can bind with another substrate for reuse.
There are two major types of molecules that modulate enzyme reaction or activity: (1) inhibitors and (2) activators.
- Inhibitors are molecules that inhibit or decrease enzymatic activity.
- Activators are molecules that stimulate or increase enzymatic activity.
Others that can affect the activity of enzymes are denaturants (chaotropic agents).
Inhibition of Enzyme Activity
Inhibition of enzymatic reactions occurs when an inhibitor binds to a site of the enzyme. This makes the enzyme not as effective as it is in forming the usual product or in performing the usual reaction.
Different Inhibition Mechanisms of Enzyme Activity |
|
|---|---|
|
|
|
Irreversible inhibition is mediated by irreversible inhibitors that interact with the enzyme via strong chemical bonds (e.g., covalent bonds). Because of this, the interaction cannot be readily reversed. The binding of the inhibitor and the enzyme leads to crucial structural changes, particularly in the active site of the enzyme. The inhibitor cannot be easily removed from the enzyme or is rather permanent.
Example: diisopropyl fluorophosphate (DIFP) interacts and thus inhibits the peptidases. trypsin and chymotrypsin. |
|
Reversible inhibition is mediated by reversible inhibitors that interact with the enzyme via weak chemical bonds (noncovalent bonds) and so the interaction can be easily reversed. It may or may not cause structural changes in the enzyme. The inhibitor can easily dissociate from the enzyme. |
|
|
|
|
|
|
|
|
Note:
|
|
Data Source: Maria Victoria Gonzaga of Biology Online
This figure below might help you understand further enzyme kinetics. Or read this guide to understand the concepts of Vmax and Km.
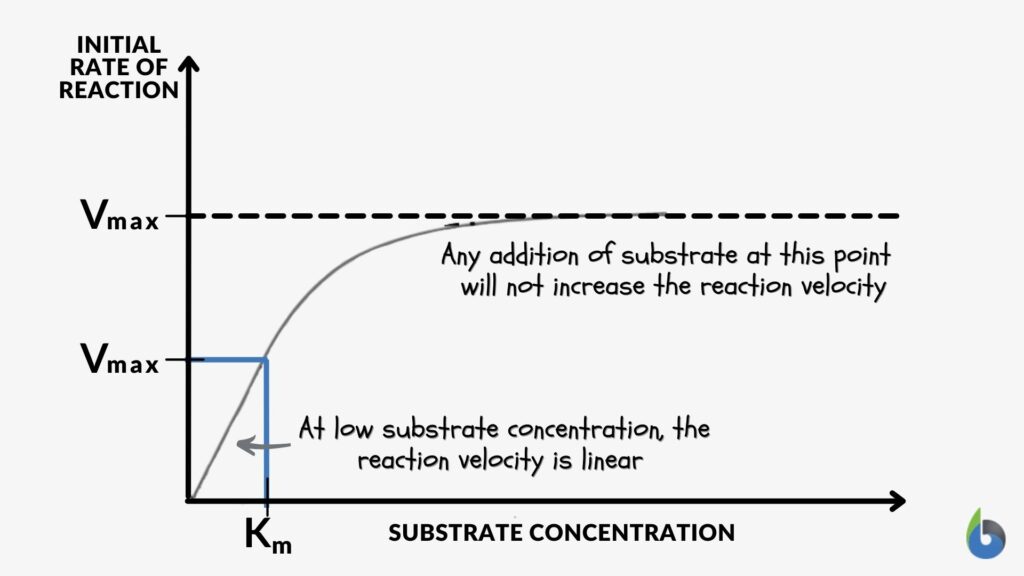 |
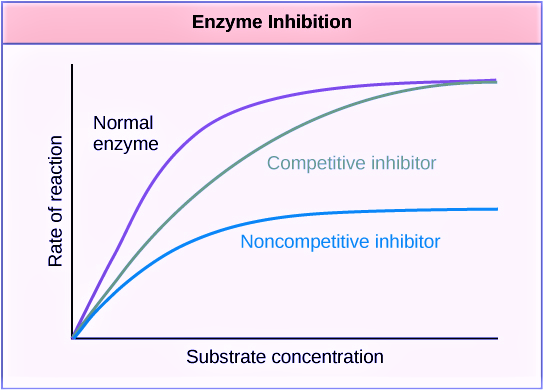 |
| Figure 4: Left: Enzyme kinetics graph: Reduced Vmax value means a reduced amount of enzymes. Image Source: Maria Victoria Gonzaga of Biology Online. Right: Enzyme Kinetics graph – enzyme inhibition. Image Credit: OpenStax College, CC BY 3.0. A low Km value means a high affinity of the enzyme to its substrate. Conversely, a high Km value means a low affinity of the enzyme to its substrate. | |
Cofactors – Metal ions and Coenzymes
Some enzymes require non-protein molecules called cofactors for their catalytic activities. The cofactor may be metal ions or coenzymes. Cofactors bind usually to the active site of the enzyme. When the cofactor is unbound, the enzyme is referred to as apoenzyme; When bound, the enzyme is referred to as a holoenzyme (however, holoenzyme also refers to an enzyme that has multiple protein subunits).
- Metal ions that serve as cofactors include iron, magnesium, manganese, cobalt, copper, zinc, and molybdenum. (Chapter 7: Catalytic Mechanisms of Enzymes – Chemistry, 2015)
- A coenzyme is a molecule that transports chemical groups (e.g., hydride ion, phosphate group, acetyl group, methyl group, etc.) from one enzyme to another. Prosthetic groups refer to the coenzymes that are tightly bound to the enzymes. Coenzymes are derived from vitamins:
- NADH, from vitamin B3 (niacinamide)
- NADPH, from vitamin B3 (niacin – carboxylic acid form)
- FMN (flavin mononucleotide), from vitamin B2 (riboflavin)
- FAD (flavin adenine dinucleotide), from vitamin B2 (riboflavin)
- TPP (thiamine diphosphate), from vitamin B1 (thymine)
- THF (tetrahydrofolate), from vitamin B9 (folic acid)
- Pyridoxyl 5′-phosphate and Pyridoxamine 5′-phosphate, from vitamin B6 (pyridoxine)
- Adenosylcobalamin, from vitamin B12 (cyanocobalamin)
- Biotin-Enzyme complex, from biotin
- Coenzyme A, from pantothenic acid
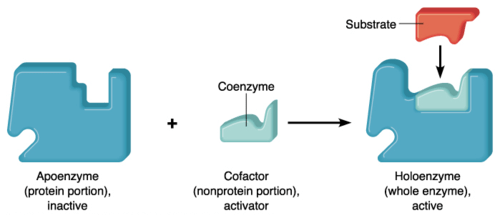
Watch this vid to understand holoenzyme, coenzyme, apoenzyme, and cofactor:
Biological Role and Function
Enzymes are biological catalysts. As catalysts, enzymes are apparently not required for a chemical reaction to happen. However, the role of enzymes in biological processes and various metabolic reactions becomes crucial when time plays a huge factor. Enzymes speed up biochemical reactions. Almost all metabolic reactions use various enzyme molecules to accelerate the conversion of substrates into products. Without such enzymes, quick biosynthesis of biomolecules is unlikely. Just take a look at the biomolecular processes associated with the Central Dogma of Life:
- DNA polymerases are enzymes that assist in DNA replication
- RNA polymerases are enzymes that assist in mRNA synthesis (transcription)
- Ribozymes (Ribosomes) are enzymes that assist in protein translation

Enzymes speed up the reaction rate about a million times faster over a process without an enzyme. But the question is, how do they do it? Enzymes have the ability to lower the activation energy required for a chemical reaction to occur. (Khan Academy, 2022)
Apart from lowering the activation energy, enzymes play a crucial role in minimizing the time required for the reaction to occur through completion. Similar to any catalyst, an enzyme would be able to speed up a chemical reaction without altering the equilibrium of a reaction. It means that a catalyst is not consumed in a reaction. Nevertheless, an enzyme differs from non-biological catalysts in being relatively more specific. Before an enzyme can catalyze a reaction, it must bind to its substrate first.
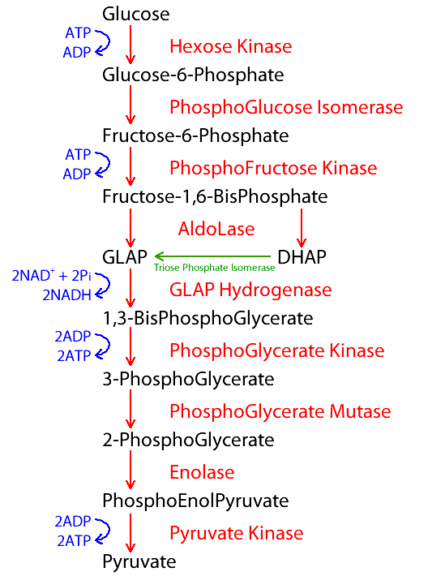
Some enzymes work in unison for the same reaction or biological process. Take glycolysis as an example where more than one enzyme is employed. Such a process employs several enzymes in a specific order, and thus, forms a distinct metabolic pathway.
Trivia!
Question: What is the fastest enzyme?
Answer: Carbonic anhydrase is believed to be the fastest enzyme found in nature so far. It has reaction rates on the order of 106/s. (Rogers et al., 2012) It catalyzes the reaction where carbon dioxide and water are converted into carbonic acid (i.e., bicarbonate and hydrogen ions): CO2 + H2O ↔ H+ + HCO3−. Thus, it helps in regulating pH (acid-base homeostasis). This enzyme is thought of as the fastest and one of the reasons it is so is because its substrates and products are relatively small molecules with low molecular weights, which accounts for their high diffusion rates. This means that this enzyme can perform as fast as the substrates have been made available by diffusion.
Partially Reviewed by: Todd Smith, PhD
Take the Quiz!
Further Reading
References
- Khan Academy. (2022). Khanacademy.org. https://www.khanacademy.org/science/ap-biology/cellular-energetics/environmental-impacts-on-enzyme-function/a/hs-enzymes-review#:~:text=Factors%20affecting%20enzyme%20activity,to%20bind%20to%20a%20substrate.
- NT Contributor. (2017, March 27). Anatomy and physiology of ageing 3: the digestive system | Nursing Times. Nursing Times. https://www.nursingtimes.net/roles/older-people-nurses-roles/anatomy-and-physiology-of-ageing-3-the-digestive-system-27-03-2017/
- Cleveland Clinic. (2021). Elevated Liver Enzymes: What Is It, Causes, Prevention & Treatment. Cleveland Clinic. https://my.clevelandclinic.org/health/symptoms/17679-elevated-liver-enzymes#:~:text=If%20you%20have%20high%20levels,also%20cause%20elevated%20liver%20enzymes.
- Fabry disease: MedlinePlus Genetics. (2022). Medlineplus.gov. https://medlineplus.gov/genetics/condition/fabry-disease/
- Rogers, D. M., Jiao, D., Pratt, L. R., & Rempe, S. B. (2012). Structural Models and Molecular Thermodynamics of Hydration of Ions and Small Molecules. Annual Reports in Computational Chemistry Volume 8, 71–127. https://doi.org/10.1016/b978-0-444-59440-2.00004-1
- Chapter 7: Catalytic Mechanisms of Enzymes – Chemistry. (2015). Wou.edu. https://wou.edu/chemistry/courses/online-chemistry-textbooks/ch450-and-ch451-biochemistry-defining-life-at-the-molecular-level/chapter-7-catalytic-mechanisms-of-enzymes/#catalytic
© Biology Online. Content provided and moderated by Biology Online Editors